It's a Long Way to the Top If You Wanna Rock 'N' Roll
What we do
Our work combines “small-scale biology” of protein molecular mechanisms and “large-scale” network biology. We seek to answer how structural and biochemical properties of ultrasensitive regulatory switches, such as GTPase conformations or protein phosphorylation, encode for simultaneous, yet precise, regulation of many different cellular processes. We approach these questions with systems biochemistry – a study of protein properties that only become apparent once the protein is studied in the context of the whole cellular network.
Specifically, we use CRISPR-based genome editing methods (e.g., prime editing) to introduce mutations that differentially perturb biochemical properties of regulatory switches. These mutations, e.g., perturb the switch kinetics of GTPases, or the negative feedback regulation of kinases. To obtain a systems-level, quantitative map of how the different mutations affect cellular functions, we use CRISPR/Cas9-based genetic interaction screens. These screens quantify the growth fitness of each point mutant tens of thousands of times, each in the context of an expression perturbation of a different human gene. Genetic interaction screens are a method for unbiased and quantitative measurement of phenotype and a resulting genetic interaction map of a mutation identifies genes involved in all cellular processes that are perturbed by that specific mutation.
There are currently two main research directions in the lab: i) on the functional multi-specificity of RAN GTPase; and ii) on the functional crosstalk between EGF and insulin signaling.
Functional multi-specificity of RAN GTPase
RAN GTPase is a model protein switch that exemplifies functional multi-specificity. By molecular function RAN is a two-state switch with two opposing regulators, switching it to GTP-bound (by RCC1, the RANGEF) and GDP-bound (by RANGAP1). In the cell, the RAN switch interacts with dozens of other proteins, and their concerted effect determines RAN’s cellular function. Consequently, RAN is the driver of nucleocytoplasmic transport, but it also has essential roles in several aspects of the cell cycle, nuclear envelope maintenance, and RNA processing. While this binding promiscuity in combination with switch ultrasensitivity enables RAN to integrate and co-regulate multiple different cellular processes it also makes it impossible to reconstitute its function by only biochemical or only genetic approaches.
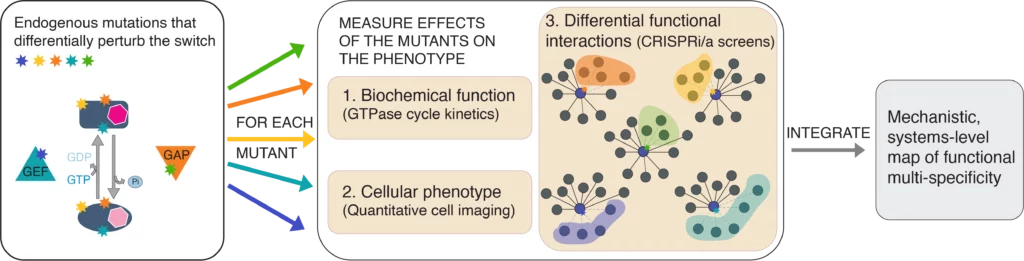
We probe the RAN switch with point mutations in RAN, GAP, or GEF, and quantify the effects of these point mutations on all the levels of RAN function: biochemical parameters, cellular localization, and systems-level biological functions. The goal is to integrate these separate levels into a single mechanistic systems wide model of RAN function.
For any questions about the RAN project contact Tina or Judith.
The functional crosstalk between EGF and insulin signalling
Canonically defined signal transduction pathways are not insulated from each other – they crosstalk by sharing components and regulating each other by positive and negative feedback. The fine-tuning of the EGF/MAPK/ERK signal transduction via negative and positive feedback loops within the pathway must be essential for its functional interactions with other pathways, such as e.g., insulin signalling. But how exactly does this complex pathway architecture of the EGF/MAPK/ERK pathway enable this crosstalk?
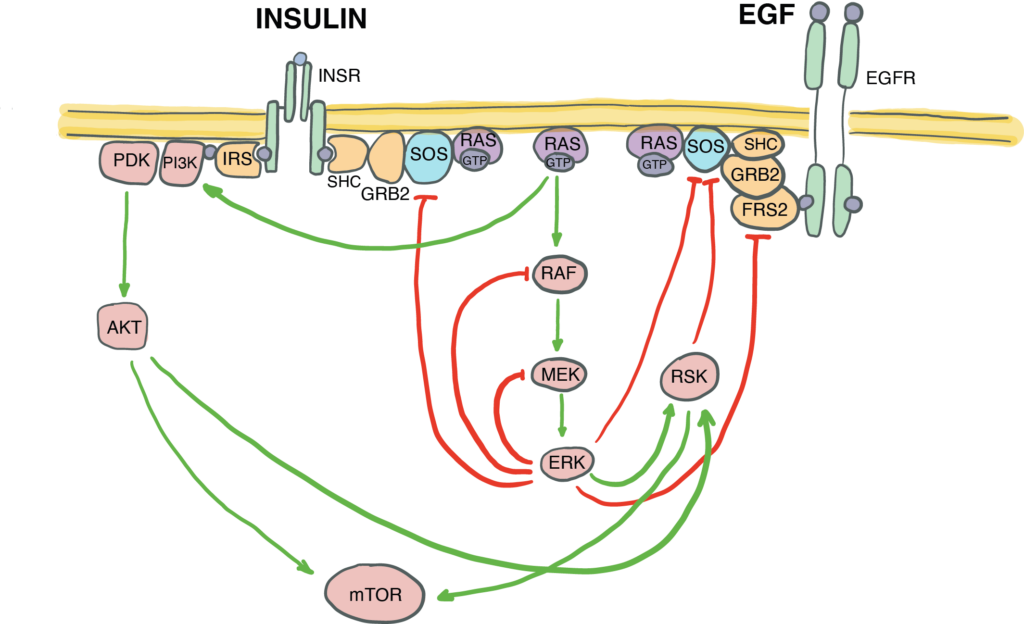
We combine immunofluorescence-based quantification and proteome wide Tandem Mass Tag Mass Spectrometry (TMT LC-MS/MS) to quantitatively map all the phosphorylation events at different time points upon stimulation of cells with EGF, insulin, or a combination of the two. We are now probing the EGF signalling pathway with mutations, and mapping individual regulatory loops with the functional crosstalk with insulin signalling.
For any questions about the MAPK/ERK project contact Tina or Ignacio.